Determining the in-plane orientation of single dyes in DNA origami
Kristina Hübner(1), Himanshu Joshi(2), Aleksei Aksimentiev(2), Fernando D. Stefani(3,4), Philip Tinnefeld(1) and Guillermo P. Acuna(5)
(1) Department of Chemistry and Center for NanoScience, LMU München, München, Germany
(2) Department of Physics, University of Illinois at Urbana−Champaign, Urbana, United States
(3) Centro de Investigaciones en Bionanociencias (CIBION), Buenos Aires, Argentina
(4) Departamento de Física, Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires, Buenos Aires, Argentina
(5) Department of Physics, University of Fribourg, Fribourg, Switzerland
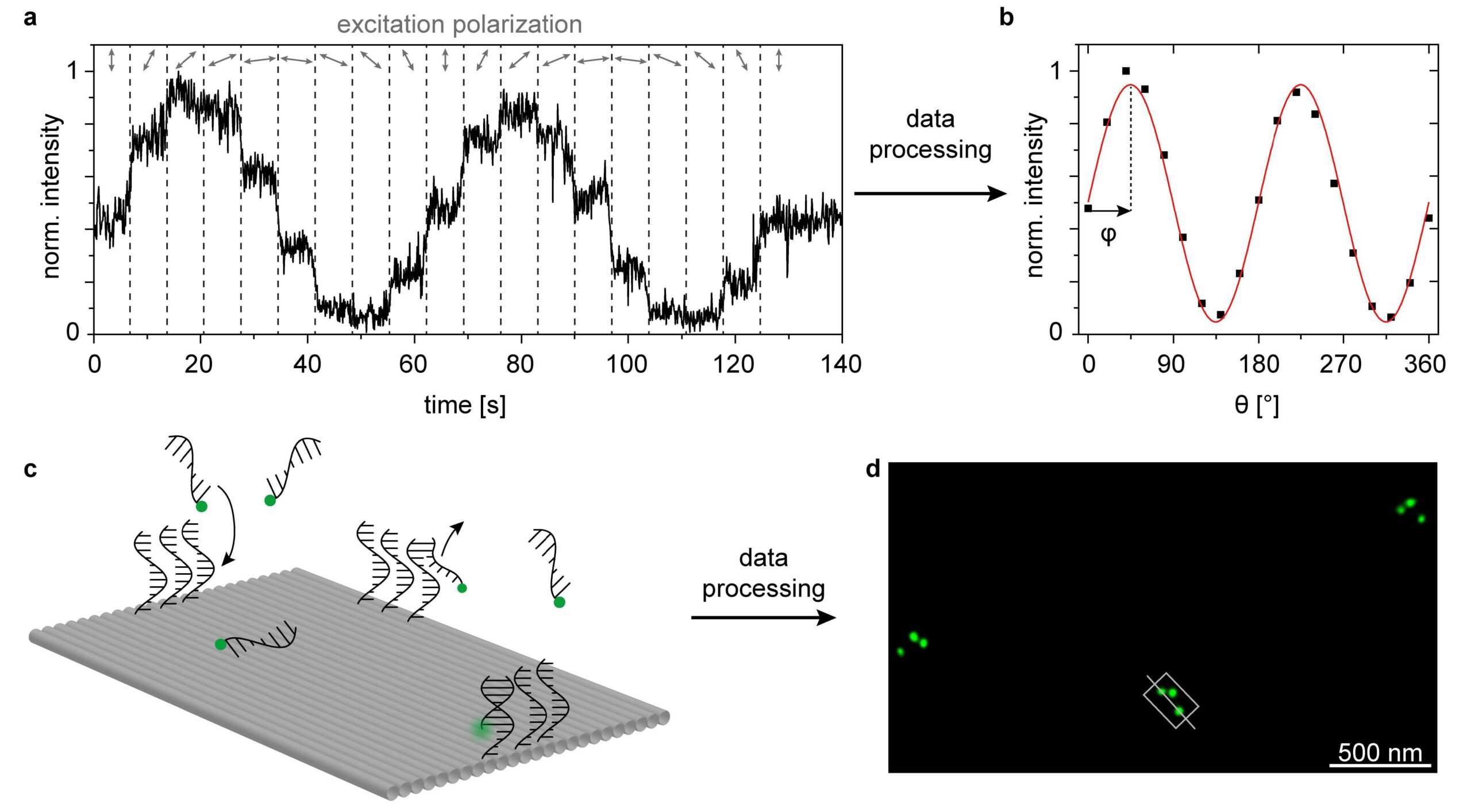
Over the last decade, DNA nanotechnology has been increasingly used to self-assemble functional nanostructures. This approach has several advantages, the most important being that different species including high quality colloidal nanoparticles and single photon emitters such as fluorophores and quantum dots can be positioned with nm precision and stoichiometric control [1,2]. In order to fully manipulate the interaction between these species, a key factor for the development of nanophotonic devices, it is necessary to not only control their relative position but also their relative orientation. Anisotropic nanoparticles, such as nanorods and triangular plates were successfully oriented within DNA origami structures [3,4]. However, the orientation of single molecules, such as fluorophores, covalently attached to DNA origami has been largely overlooked, let alone controlled. In this work [5], we bring together a method to determine the orientation of single fluorophores attached to DNA origami structures based on two independent measurements carried out in a standard wide-field fluorescence microscope. First, the orientation of the absorption transition dipole of the molecule is determined through a polarization-resolved excitation measurement (figure 1a and b). Second, the orientation of the DNA origami structure is obtained from a DNA-PAINT nanoscopy measurement (figure 1c and d). Our results show that covalently attached fluorophores can adopt different orientations on a DNA-origami depending on the immediate molecular environment. In addition, we perform all-atom molecular dynamics simulations that provide insight into the fluorophore´s binding modes. We consider that this work is a first step towards the orientation control of single fluorophores in DNA origami structures, that will enable the development of more efficient and reproducible selfassembled nanophotonic devices.
1 – (a) Polarization-resolved excitation measurements. (b) Mean fluorescence intensity vs θ to obtain the in-plane orientation (φ) of the excitation transition dipole moment. (c) DNA-PAINT nanoscopy measurements. Sketch of the binding and unbinding of the imager strands labelled with a single ATTO 542 onto the DNA-PAINT binding sites. (d) Super resolved DNA-PAINT image corresponding to three DNA origami structures showing the asymmetric triangular pattern. Based on this image, the orientation in 2D as well as the binding geometry (upright or upside down) of each DNA origami (grey rectangle) can be extracted (greyline).
References:
[1] Kuzyk, A. et al., ACS Photonics, 5, 1151 (2018).
[2] Hübner, K. et al., Nano Lett., 19, 6629 (2019).
[3] Pal, S. A. et al., J. Am. Chem. Soc., 133, 17606 (2011).
[4] Zhan, P., Angew. Chemie Int. Ed., 57, 2846 (2018).
[5] Hübner, K. et al., Nano Lett., 15, 5109 (2021).
Acknowledgment: This work was supported from the Swiss National Science Foundation (200021_184687) and from the ITN SuperCol funded by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement 860914
© 2025 · Biophotonics4Future
Leibniz Institute of Photonic Technology
Albert-Einstein-Str. 9
07745 Jena | Germany
www.leibniz-ipht.de